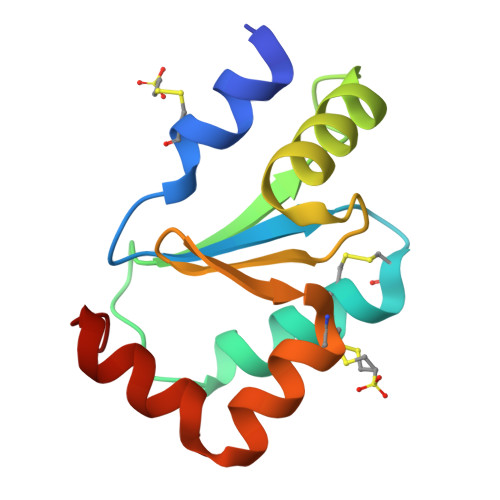
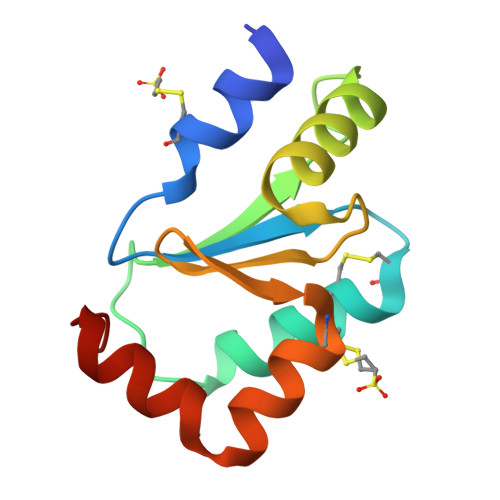
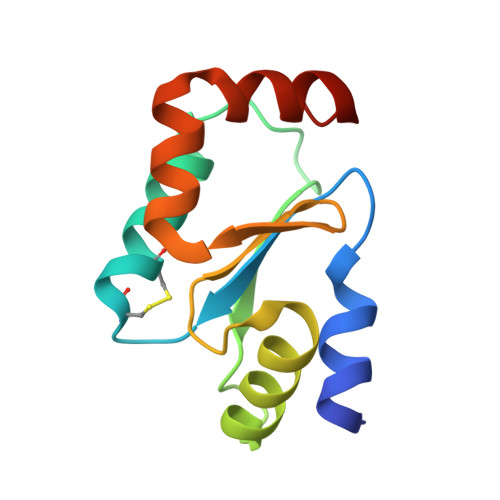
Cysteine Specific Targeting of the Functionally Distinct Peroxiredoxin and Glutaredoxin Proteins by the Investigational Disulfide BNP7787.
Parker, A.R., Petluru, P.N., Nienaber, V.L., Badger, J., Leverett, B.D., Jair, K., Sridhar, V., Logan, C., Ayala, P.Y., Kochat, H., Hausheer, F.H.(2015) Molecules 20: 4928-4950
- PubMed: 25793542
- DOI: https://doi.org/10.3390/molecules20034928
- Primary Citation of Related Structures:
4RQR, 4RQX - PubMed Abstract:
Glutaredoxin (Grx), peroxiredoxin (Prx), and thioredoxin (Trx) are redoxin family proteins that catalyze different types of chemical reactions that impact cell growth and survival through functionally distinct intracellular pathways. Much research is focused on understanding the roles of these redoxin proteins in the development and/or progression of human diseases. Grx and Prx are overexpressed in human cancers, including human lung cancers. BNP7787 is a novel investigational agent that has been evaluated in previous clinical studies, including non-small cell lung cancer (NSCLC) studies. Herein, data from activity assays, mass spectrometry analyses, and X-ray crystallographic studies indicate that BNP7787 forms mixed disulfides with select cysteine residues on Grx and Prx and modulates their function. Studies of interactions between BNP7787 and Trx have been conducted and reported separately. Despite the fact that Trx, Grx, and Prx are functionally distinct proteins that impact oxidative stress, cell proliferation and disease processes through different intracellular pathways, BNP7787 can modify each protein and appears to modulate function through mechanisms that are unique to each target protein. Tumor cells are often genomically heterogeneous containing subpopulations of cancer cells that often express different tumor-promoting proteins or that have multiple dysregulated signaling pathways modulating cell proliferation and drug resistance. A multi-targeted agent that simultaneously modulates activity of proteins important in mediating cell proliferation by functionally distinct intracellular pathways could have many potentially useful therapeutic applications.
Organizational Affiliation:
BioNumerik Pharmaceuticals, Inc., 8122 Datapoint Drive, Ste. 1250, San Antonio, TX 78229, USA.